| Host: | Rabbit |
| Applications: | WB/IHC/IF/ELISA |
| Reactivity: | Human/Mouse |
| Note: | STRICTLY FOR FURTHER SCIENTIFIC RESEARCH USE ONLY (RUO). MUST NOT TO BE USED IN DIAGNOSTIC OR THERAPEUTIC APPLICATIONS. |
| Short Description: | Rabbit polyclonal antibody anti-Phospho-Replication protein A 32 kDa subunit-Thr21 (10-59 aa) is suitable for use in Western Blot, Immunohistochemistry, Immunofluorescence and ELISA research applications. |
| Clonality: | Polyclonal |
| Conjugation: | Unconjugated |
| Isotype: | IgG |
| Formulation: | Liquid in PBS containing 50% Glycerol, 0.5% BSA and 0.02% Sodium Azide. |
| Purification: | The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen. |
| Concentration: | 1 mg/mL |
| Dilution Range: | WB 1:500-1:2000IHC 1:100-1:300ELISA 1:10000IF 1:50-200 |
| Storage Instruction: | Store at-20°C for up to 1 year from the date of receipt, and avoid repeat freeze-thaw cycles. |
| Gene Symbol: | RPA2 |
| Gene ID: | 6118 |
| Uniprot ID: | RFA2_HUMAN |
| Immunogen Region: | 10-59 aa |
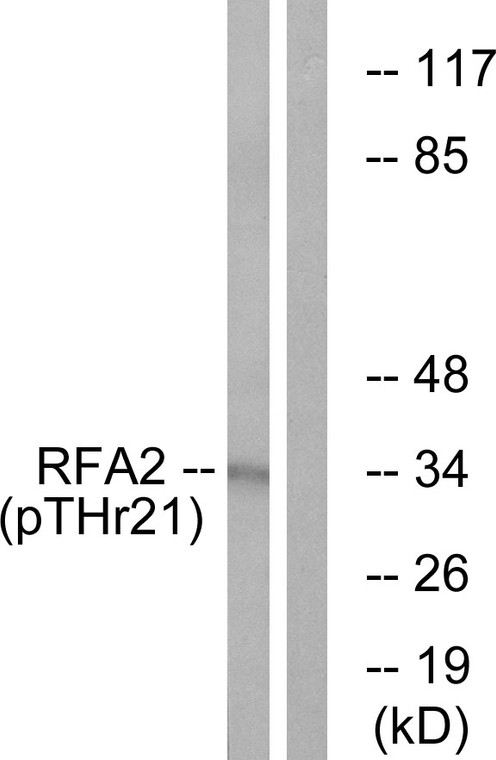
| Specificity: | Phospho-RPA32 (T21) Polyclonal Antibody detects endogenous levels of RPA32 protein only when phosphorylated at T21. |
| Immunogen: | The antiserum was produced against synthesized peptide derived from the human RFA2 around the phosphorylation site of Thr21 at the amino acid range 10-59 |
| Function | As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism. Thereby, it plays an essential role both in DNA replication and the cellular response to DNA damage. In the cellular response to DNA damage, the RPA complex controls DNA repair and DNA damage checkpoint activation. Through recruitment of ATRIP activates the ATR kinase a master regulator of the DNA damage response. It is required for the recruitment of the DNA double-strand break repair factors RAD51 and RAD52 to chromatin in response to DNA damage. Also recruits to sites of DNA damage proteins like XPA and XPG that are involved in nucleotide excision repair and is required for this mechanism of DNA repair. Also plays a role in base excision repair (BER) probably through interaction with UNG. Also recruits SMARCAL1/HARP, which is involved in replication fork restart, to sites of DNA damage. May also play a role in telomere maintenance. |
| Protein Name | Replication Protein A 32 Kda SubunitRp-A P32Replication Factor A Protein 2Rf-A Protein 2Replication Protein A 34 Kda SubunitRp-A P34 |
| Database Links | Reactome: R-HSA-110312Reactome: R-HSA-110314Reactome: R-HSA-110320Reactome: R-HSA-174437Reactome: R-HSA-176187Reactome: R-HSA-3371453Reactome: R-HSA-3371511Reactome: R-HSA-5358565Reactome: R-HSA-5358606Reactome: R-HSA-5651801Reactome: R-HSA-5655862Reactome: R-HSA-5656121Reactome: R-HSA-5656169Reactome: R-HSA-5685938Reactome: R-HSA-5685942Reactome: R-HSA-5693607Reactome: R-HSA-5693616Reactome: R-HSA-5696395Reactome: R-HSA-5696397Reactome: R-HSA-5696400Reactome: R-HSA-6782135Reactome: R-HSA-6782210Reactome: R-HSA-6783310Reactome: R-HSA-6804756Reactome: R-HSA-68962Reactome: R-HSA-69166Reactome: R-HSA-69473Reactome: R-HSA-912446Reactome: R-HSA-9709570 |
| Cellular Localisation | NucleusPml BodyRedistributes To Discrete Nuclear Foci Upon Dna Damage In An Atr-Dependent Manner |
| Alternative Antibody Names | Anti-Replication Protein A 32 Kda Subunit antibodyAnti-Rp-A P32 antibodyAnti-Replication Factor A Protein 2 antibodyAnti-Rf-A Protein 2 antibodyAnti-Replication Protein A 34 Kda Subunit antibodyAnti-Rp-A P34 antibodyAnti-RPA2 antibodyAnti-REPA2 antibodyAnti-RPA32 antibodyAnti-RPA34 antibody |
Information sourced from Uniprot.org
12 months for antibodies. 6 months for ELISA Kits. Please see website T&Cs for further guidance